Bartonella and a few other low-yield microbes pose significant challenges to conventional diagnostic testing. Healthcare providers who are familiar with illnesses caused by these pathogens will often clinically diagnose patients based on their exposure risks, travel history, and symptoms, but without laboratory confirmation. Why?
Despite recent innovations, developing a diagnostic test that is 100% reliable for confirmation of Bartonella species infection is nearly impossible. There is almost always a possibility of false positive or false negative results due to test design, technological challenges, and other implicit factors. These testing challenges are especially relevant for infections caused by elusive flea- and tick-borne microbes, like Bartonella species, that may lead to chronic illnesses that start with non-specific symptoms (fever, headache, swollen glands, aches and pains).
There are many direct and indirect laboratory test options for bartonellosis:
- Direct Detection: Confirms the presence of a pathogen. DNA testing (aka nucleic acid detection) targets a specific sequence of the microbe’s DNA using detection methods like PCR, FISH, etc. Other types of direct detection methods include antigen and/or protein capture methods, bacterial isolation or plate culture, and microscopy.
- Indirect Detection: Determines whether there has been prior or recent exposure to a pathogen. Serology tests detect IgM or IgG antibodies generated by the host’s immune system response that are specific to the target pathogen.
Numerous offerings often lead providers and patients to ask us, “Which test should I order?” or “Which test is best?” There is no clear answer to this question, unfortunately. Head-to-head studies comparing test performances in clearly defined patient populations are necessary to determine clinical utility. Further, research suggests that one test method may not be adequate for all cases.
A 2011 study co-authored by Galaxy scientists documented Bartonella species infection in rheumatology patients using DNA and antibody testing. Importantly, the study found that there was minimal overlap between patients who tested positive using both direct and indirect test methods. Patients were more likely to be positive for either Bartonella species DNA or antibodies against Bartonella species. Other studies in different patient groups and high-exposure groups confirmed these findings.
For this reason, infectious disease experts recommend a combination of both direct and indirect test methods to maximize the diagnostic data available to the provider.
What makes Bartonella Detection so Challenging?
The ways these pathogens behave in the host’s body and interact with the host’s immune response often make serial or repeat testing necessary before laboratory confirmation is possible. Bartonella species pursue stealth infection strategies that make detection of DNA or antibodies very challenging, as discussed below.
Immune Evasive Mechanisms
Bartonella species are intracellular pathogens that can infect multiple cell types, cycling in and out of the bloodstream and travelling to various parts of the body via the vascular system. The primary niche for intracellular infection is the endothelial cells that line the vascular system; research suggests that invasion of bone marrow progenitor cells and skin cells (collagen) occurs as well.
Bartonella can also form biofilms. Intracellular bacteria are found at very low concentrations in blood, urine, tissue and other specimens, making DNA detection very challenging.
Slow-Growing, Low-Yield Infections
Bartonella species replicate very slowly, even when growth conditions are ideal. The doubling time can be 22-24 hours; by contrast, E. coli may take only 20-30 minutes. In other words, the infectious dose given from the feces of a flea or the bite of a tick is likely low, and the concentration of bacteria within the blood or tissue remains low throughout the course of the infection.
Host Immune Response
The host’s immune response leads to the development of antibodies that help protect the body from current or future infection. Bartonella species infection leads to delays in antibody production due to their slow growth rates and ability to “hide” in multiple areas of the body. Delayed antibody response against Bartonella species infection was first observed in HIV patients and may not be detectable for 6 weeks or more after acute infection in people with competent immune systems. Research further suggests that some patients do not show antibodies against Bartonella species until after the initiation of antibiotic therapy (aka seroconversion).
Case reports have also implicated Bartonella species in immune system dysfunction. This means that the bacteria could be negatively impacting the immune response by directly affecting white blood cell production. The mechanisms for this require more research to be fully understood.
Test Improvements
Novel, or improved, direct detection methods are needed to overcome the various challenges described above and to ultimately reduce the amount of testing necessary to confirm Bartonella species infection. Galaxy scientists are focused on developing techniques that enrich or manipulate the sample to increase the sensitivity of direct detection methods.
Sample enrichment or manipulation is necessary for low-yield infections because PCR assays have a limit of detection (LOD). If the amount of target DNA that is present is not sufficient, it will not be detected. Further, the small target signal may be imperceptible due to overwhelming background noise from inhibitors present in the sample.
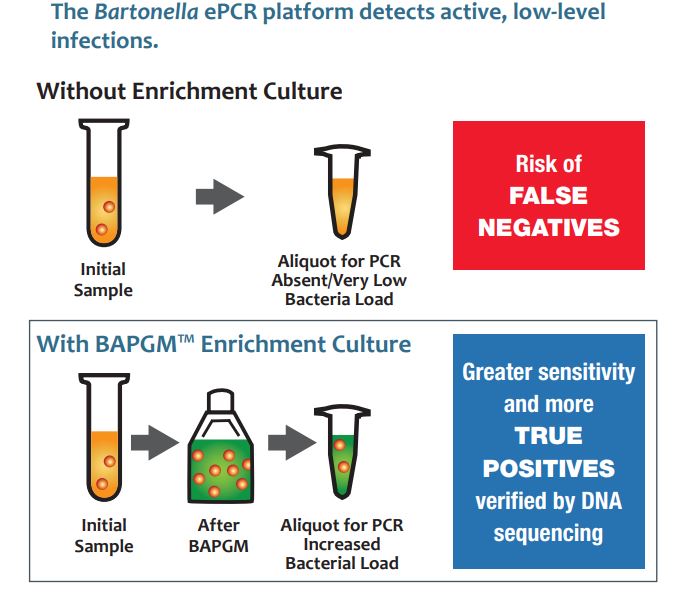
Sample enrichment in our proprietary BAPGM™ liquid culture was the first technique we employed to increase the amount of bacterial target for detection by qPCR. We called this BAPGM™ enrichment platform Bartonella ePCR™.
This testing platform combined a week-long sample enrichment in our proprietary BAPGM™ liquid culture with genus-level PCR to cover a broad range of Bartonella species. BAPGM™ culture “enriches” the sample by growing the few bacteria present in a given specimen up to detectable levels for confirmation by PCR. With this approach, scientists at Galaxy, NCSU and other research institutions around the world have been able to drive research findings revealing the clinical importance of Bartonella infection across a broad array of symptoms and patient groups.
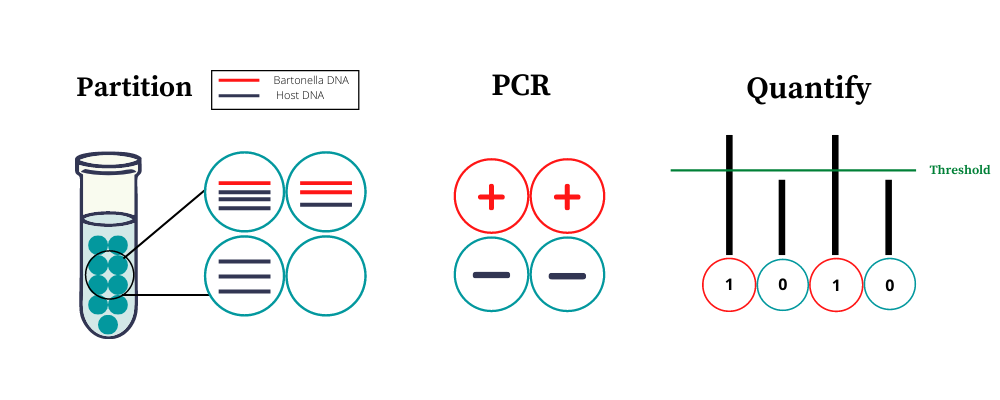
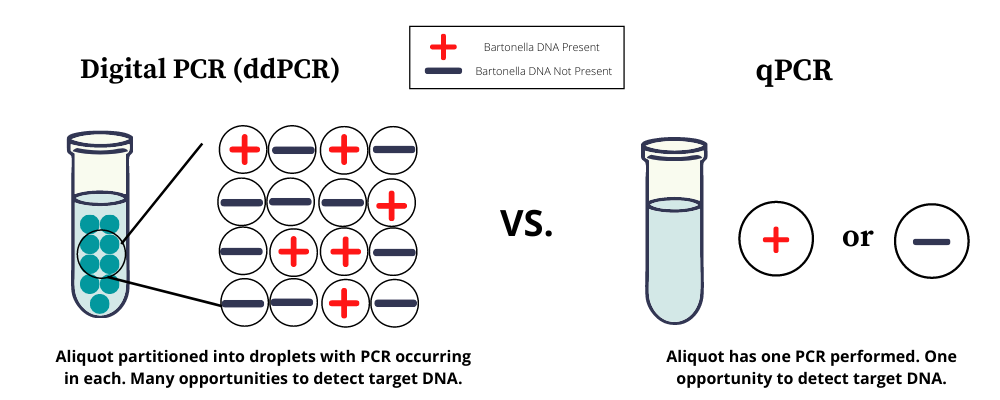
Galaxy and NCSU scientists recently co-authored a publication that explains how digital PCR (or ddPCR) improves upon standard qPCR with and without BAPGM™ enrichment. Instead of performing a single PCR reaction on a sample, ddPCR technology partitions each small PCR sample into 20,000 droplets and performs a PCR reaction on each droplet, which reduces the noise of inhibitors like host DNA.
Normally, testing for DNA evidence of a few copies of bacteria in a PCR sample is like trying to find a needle (target DNA) in a haystack (inhibitors). You can look for the needle in a giant stack of hay with limited success, or you can spread the hay out across a field and look for the needle systematically in each square inch.
Although the launch of ddPCR itself will certainly lead to huge strides in improving direct detection of Bartonella and other low-yield infections, the preliminary data from this study suggest that it is still necessary to combine ddPCR with BAPGM™ enrichment in order to maximize molecular sensitivity. Further development is required to determine clinical sensitivity in different patient groups and at different stages and levels of infection in comparison to IFA serology.
Bartonella Digital ePCR™ is now available!
Earlier this year, we announced the launch of Bartonella Digital ePCR™. This new and improved test platform replaces qPCR with the more sensitive ddPCR (or Digital PCR) technology described above.
References
HIV.gov. (2013). Guidelines for the Prevention and Treatment of Opportunistic Infections in Adults and Adolescents with HIV: Bartonellosis. Retrieved from: https://clinicalinfo.hiv.gov/en/guidelines/adult-and-adolescent-opportunistic-infection/bartonellosis-0?view=full
Westling, K. et al. (2008). Bartonella henselae antibodies after cat bite. Emerging Infectious Diseases, 14(12), 1943-1944. 10.3201/eid1412.080002 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2634615/
Kaufman, D. L. et al. (2017). Neurological and immunological dysfunction in two patients with Bartonella henselae bacteremia. Clinical Case Reports, 5(6), 931-935. 10.1002/ccr3.077 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5458018/
Maggi, R. G. et al. (2020). Development and validation of a droplet digital PCR assay for the detection and quantification of Bartonella species within human clinical samples. Journal of Microbiological Methods, 176, 106022. 10.1016/j.mimet.2020.106022 https://pubmed.ncbi.nlm.nih.gov/32795640/
Maggi, R.G. et al. (2011). Bartonella spp. bacteremia in high-risk immunocompetent patients. Diagnostic microbiology and infectious disease, 71(4), pp.430-437. https://pubmed.ncbi.nlm.nih.gov/21996096/