On June 2, 2020, Nextstrain clarified the clade naming for SARS-CoV-2, the virus that causes COVID-19. This brings up the question: What do the terms “subspecies”, “strain” and “clade” mean, and why does it matter to you?
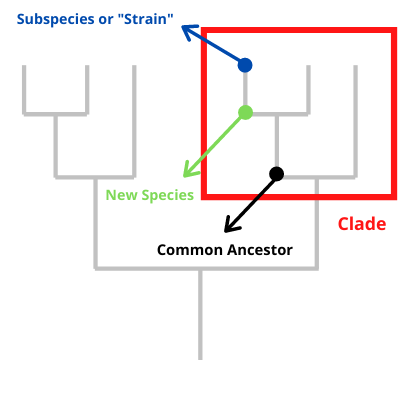
In taxonomy (the science of naming and classifying biology), the word “clade” isn’t used only to divide a species. It can be used at a level from kingdom down to genus as well to identify a division below an official place where the tree of life divides. If you’ve seen someone put up a picture of the branching tree of taxonomy and then circle a few branches, what they are doing with that circle is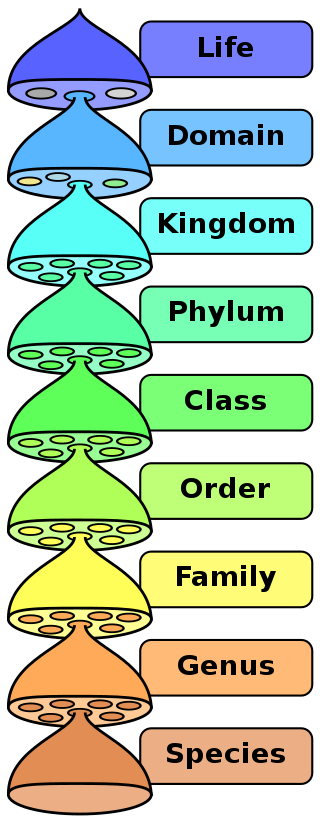 often defining a clade.
often defining a clade.
Most frequently when clades are discussed in the news, they are a division of a species or a sub-species. The genetic variance they represent isn’t enough to rise to the level of division of species, but it is worth noting by whoever is talking about it. However, this kind of division could also be called a “strain.” Why is one word used rather than the other?
When to use the word “strain” and when to use the word “clade” to describe a division below the species level isn’t precisely defined. For viruses and bacteria, both of which are characterized by non-sexual reproduction, “strain” tends to be used to refer to a genetic change that causes a change in how the species interacts with the environment, and in particular how a pathogen interacts with its host. In contrast, “clade” tends to be used for a genetic change that has no meaningful change.
For animals, “strain” tends to be used for a variation that is interacting with other variations in sexual reproduction and “clade” tends to be used when a group of individuals have genetic uniformity in the genetic variation in question.
Advances in DNA detection and sequencing techniques have allowed researchers to quickly identify and characterize emerging genetic variants of viruses and bacteria as they are found in animal and human samples. The most relevant example of this is the novel coronavirus responsible for the ongoing COVID-19 pandemic.
SARS-CoV-2
At the species level, if a clade has been identified, why not call it a strain or a new species? Generally this is because the variation isn’t meaningful enough for it to be a new species. For example, SARS-COV-2 as defined by the International Committee on Taxonomy of Viruses (ICTV) is a subspecies of severe acute respiratory syndrome-relative coronavirus. That species is identified by five genetic sequences in the genome of the virus. So long as those five genetic sequences remain consistent, any variation would be a sub-species, a strain, or a clade.
In the case of SARS-CoV-2, clades are very interesting because they can be used to track how the virus bounced around the world, entering and re-entering various regions. The variations emerge one from another, and that information allows the added element of time to be added to the tracking.
This is important from a public health standpoint because clades that are specific to a certain region can give epidemiologists insight into the extent of community-spread disease and help with contact tracing.
There is some discussion that there may be two strains of SARS-CoV-2. Some clinicians feel they have seen a difference in disease severity between what are currently called clades. A small study did not support this idea, but further research remains to be done.
Bartonella henselae
Within the Bartonella genus, several subspecies, strains and clades have been identified. For example, Bartonella vinsonii is a species commonly found in dogs that has been divided into subspecies. The subspecies berkhoffii has further been broken down into genotypes (or clades) I, II and III.
Bartonella henselae has a strain known as Houston-1 that has 9 genetic copies of a gene segment that encodes transcriptional regulatory proteins. Recent research has found that these copies of the gene interact in a way that regulates the production of biofilms and causes this strain to have an enhanced biofilm production. This is the kind of meaningful change to how the strain interacts with its environment that may someday cause it to be changed from a strain to a subspecies.
Conclusion
Recent discussion of SARS-CoV-2 clades has raised questions and concerns for the public. Exactly why a scientist might call something a strain versus a clade is not entirely defined, even for scientists.
References
Hamzelou, J. (2020). Coronavirus: Are there two strains and is one more deadly? NewScientist.com Available at: https://www.newscientist.com/article/2236544-coronavirus-are-there-two-strains-and-is-one-more-deadly/
Okaro, U. et al. (2020). A non-coding RNA controls transcription of a gene encoding a DNA binding protein that modulates biofilm development in Bartonella henselae [online pre-print]. Microbial Pathogenesis, 104272. doi:10.1016/j.micpath.2020.104272 https://pubmed.ncbi.nlm.nih.gov/32464301/?from_term=bartonella&from_sort=date&from_pos=1

